4. The advantages and disadvantages of LD
The merit of LD
One advantage is that if we have limited genetic data which do not contain the causal SNPs for the disease trait of interest, we can obtain significant results for SNPs neighboring those causal SNPs and narrow down the “SNP causal zone”. This allows subsequent research to focus more on collecting data in this area to determine the causal SNP.
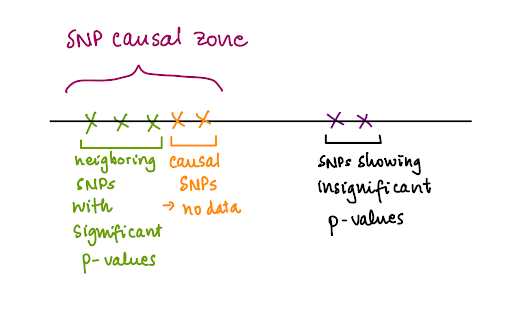
…But the problems are…
Problem 1: Difficult to figure out the causal SNPs
If a lot of the SNPs are significant, it will be hard to determine the exact SNP(s) that cause the disease to zoom in on them.
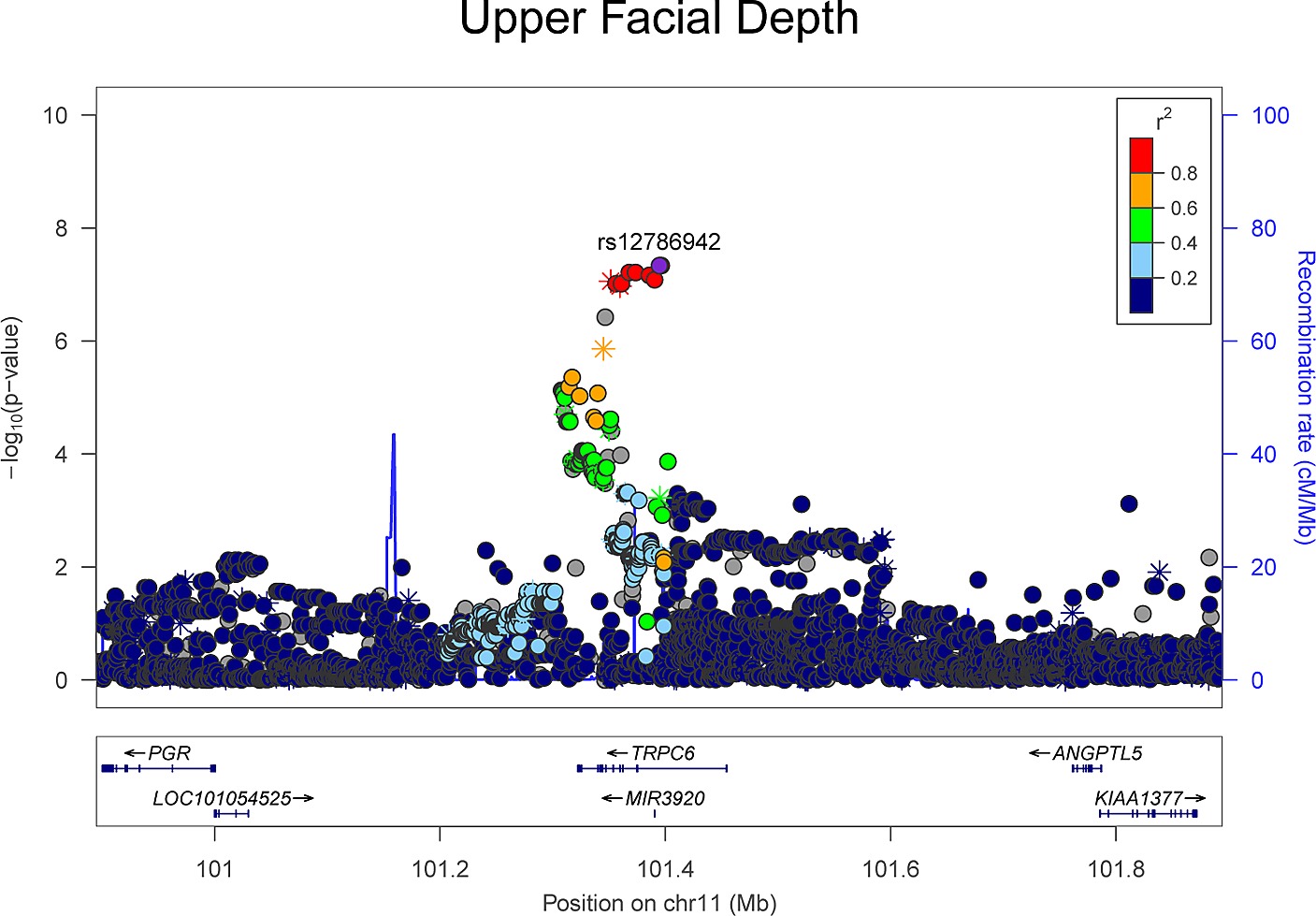
In the plot above, the purple SNP is the causal SNP, and the SNPs neighboring to it (colored in red) are highly correlated with the causal SNP. Therefore, in GWAS, although the purple SNP should be the only significant SNP, the purple- and the red-colored SNPs are likely to be found significant as well.
Problem 2: Hard to determine the p-value threshold in Multiple Testing
The Bonferroni Correction and Simulation-based Approach have been commonly used to correct for Type 1 Errors arising from Multiple Testing. However, due to their limitations when dealing with the correlation among SNPs and extensive resource.
Therefore, in our project, we aim to explore the PLIS method, which can potentially account for the correlation among SNPs using HMM and require much less resources than the Simulation-based Approach. In the next section, we aim to explain what the idea behind HMM is, and how it is used in GWAS.